Rules of multivalency-biased affinity choice
In nature, molecules typically coevolve inside interconnected, multicomponent assemblies, ensuing within the collective improvement of interfaces. As an analogy, we envisioned using a target-tailored multimerization scaffold to evolve artificial polymers towards an oligomeric goal, yielding multivalent binders with cooperative binding behaviour inside a designed multivalent meeting. Many clinically related goal proteins exist as multimeric complexes, supporting the applying of target-tailored evolution to determine new binders. The evolutionary-conserved trimeric construction of viral capsid glycoprotein complexes is emblematic of many human pathogens akin to retroviruses32, coronaviruses33 and orthomyxoviruses34. As a goal for the event of our multivalent choice methodology, we opted to make use of the trimeric SARS-CoV-2 spike protein, arguably one of the consultant members of viral class I fusion proteins (Fig. 1a).
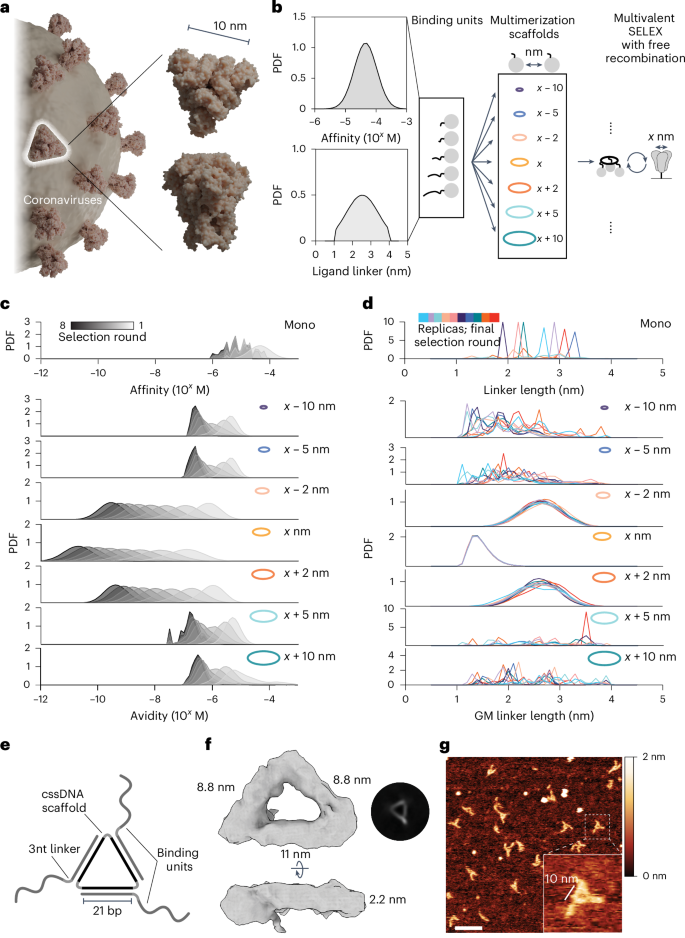
a, Giant trimeric glycoprotein complexes are attribute of many human pathogens as exemplified by SARS-CoV-2 spike protein. Determine created in Blender (https://www.blender.org), utilizing PDB 3JCL as a spike mannequin. b, Schematic for Gillespie simulation of multivalent choice towards a homotrimeric goal. A random library of binding models with various spatial tolerances for simultaneous goal engagement and a set of multimerization scaffolds with growing interligand spacing had been examined. The preliminary likelihood density perform (PDF) of affinities (prime) and linkers (backside) for the binding unit library is given. c, Simulation outcomes of multivalent choice for libraries ready with completely different multivalent scaffolds present that matching scaffold geometry facilitates the choice of assemblies with the best avidity. All Gillespie simulations had been carried out in 10 replicates (n = 10); outcomes are proven as common PDFs. d, The PDF of linkers within the binding unit library within the closing spherical of multivalent choice is decided by the scaffold geometry. GM, geometric imply. e, Schematic illustration of MEDUSA’s molecular structure. f, Cryo-EM 3D class-average electron density of a designed MEDUSA. The respective Fourier shell correlation evaluation is supplied in Supplementary Fig. 4. g, Consultant atomic pressure microscopy picture of MEDUSA. Scale bar, 20 nm.
The efficiency of multivalent binders depends closely on the scaffold becoming a member of the ligands as a result of this determines their orientation and general structural flexibility24. Earlier modelling outcomes counsel that maximal binding affinity might be achieved when the binder’s core is inflexible and matches the goal’s measurement, and the linkers have a median end-to-end distance barely longer than the space between the binder’s core and cognate binding epitopes15. Constructing on these outcomes, we developed a multiscale computational framework for simulating multivalent SELEX (Supplementary Fig. 1) via modelling stochastic choice dynamics35. Simulations used a hard and fast triangular goal with dimensions derived from the spike protein (marked as x) to look at choices carried out with varied scaffold geometries (Fig. 1b) and explored how binding affinities and linker lengths developed beneath completely different spatial constraints.
Within the multivalent SELEX course of, ligand and linker distributions adapt over successive rounds of choice, yielding one of the best avidities when the scaffold geometry intently aligns with the goal (Fig. 1c). Minor deviations in scaffold measurement (for instance, <x − 2 nm or >x + 2 nm) shortly restrict avidities to the micromolar vary, demonstrating the important position of scaffold geometry in enabling efficient multivalent binding. Scaffold geometry immediately influences linker size choice (Fig. 1d): when the scaffold geometry matches the goal geometry, libraries converge towards minimal linker lengths that facilitate multivalent engagement by exact alignment with binding epitopes. In distinction, monovalent libraries exhibit a random distribution of linker lengths after choice as a result of they don’t seem to be subjected to spatial constraints. Scaffolds deviating barely from the optimum geometry nonetheless help multivalent binding by choosing longer linkers. Nevertheless, scaffolds exterior the spatial tolerance of the ligand library fail to favour multivalent interactions. In these circumstances, choice reverts to a monovalent-like regime, with out selective strain on linker size. These findings underscore the importance of scaffold geometry in figuring out spatial constraints, offering predictive design ideas for optimizing ligand choice in multivalent methods.
Based mostly on simulation outcomes, we established a number of design ideas for the scaffold and binding unit library. First, the scaffold geometry ought to intently match the goal’s measurement and form to maximise the contribution of avidity. Second, for matching scaffolds, linkers ought to be of minimal size to not disrupt the benefits of a geometrical match. We explored a number of potential scaffold buildings that might fulfill these standards and permit for the show of binding models at ~10 nm pairwise distances inside a trivalent framework, reflecting the general dimensions and rotational symmetry of SARS-CoV-2 spike protein (Supplementary Fig. 2a,b). We settled on a cyclic single-stranded DNA (cssDNA) scaffold as a result of its nuclease stability, measurement and ease of preparation (Supplementary Figs. 2c and 3). The structure of MEDUSA emerges from the three 21-nucleotide binding unit-hybridization areas, separated by 2 T hinges inside cssDNA (Fig. 1e–g). This configuration permits the binding models to be positioned at each second flip of the DNA, displaying them on the identical face of the scaffold. Following the second design precept, we opted to maintain the linkers brief, inserting 3 nucleotides (~1.6 nm (ref. 36)) between the scaffold-hybridization area and the binding area.
Multivalent assemblies of functionalized nucleic acids
Moreover structural programmability, a serious benefit of DNA as scaffold is its compatibility with quite a few evolvable polymers, akin to DNA, RNA9,10, gradual off-rate modified aptamers37, extremely side-chain-functionalized nucleic acid polymers38, PNA39 and acyclic l-threoninol nucleic acid40. This compatibility makes it doable to assemble hybrid nanostructures that merge the versatile design potential of DNA with the chemical range of proteins and past38,41. We mix the cssDNA scaffold with functionalized nucleic acid polymer (FNAP) binding models, which collectively create the MEDUSA for affinity evolution of hybrid multivalent concentrating on nanomaterials. FNAP is a base-modified nucleic acid polymer, produced through DNA-ligase-mediated polymerization of trinucleotide constructing blocks on the DNA template. Within the FNAP library design, we partially sacrifice the density of side-chain functionalization in favour of sequence range, permitting the library to pattern a broader vary of secondary buildings of the polymer. Construction–exercise relationship research present the rationale for this alternative as a result of typically solely few aspect chains are important for goal binding38. A handy property of MEDUSA is its glorious nuclease resistance, so important for purposes in organic fluids, ensured by the cyclic scaffold and enzyme-inhibiting nucleotide modifications on the 5′ place42,43.
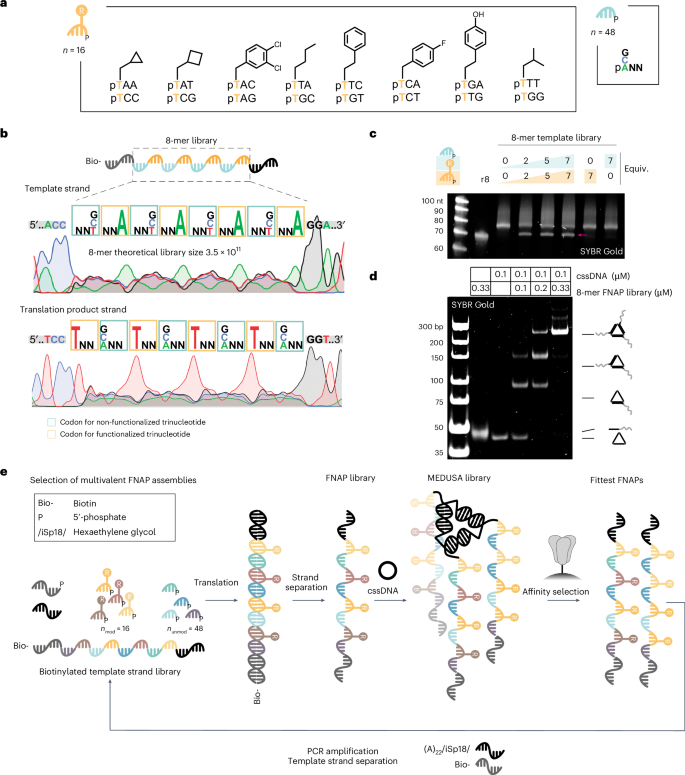
We constructed trinucleotide libraries of 16 side-chain-functionalized trinucleotides encoding 8 completely different aspect chains and 48 non-functionalized trinucleotides. Based mostly on earlier reviews44,45, we restricted side-chain range to hydrophobic residues, together with people who mimic aspect chains of proteinogenic amino acids and people not present in pure proteins (Fig. 2a, Supplementary Figs. 5 and 6, and Supplementary Desk 1). The pertinent template library options an structure that intersperses low-sequence range and side-chain-functionalized trinucleotide areas with high-diversity, non-functionalized trinucleotide areas. The previous expands the chemical range, whereas the latter enhances the polymer’s conformational range (Fig. 2b). To reduce interference between binding models inside MEDUSA, the library size was saved shorter than typical SELEX libraries (40 nt) and the typical size of reported spike-binding aptamers (Supplementary Fig. 7a). To estimate the interpretation effectivity, we ready corresponding biotinylated template libraries of 8-mer and 12-mer lengths (Supplementary Fig. 7b) and noticed a plateau already at 2 equiv. of trinucleotide libraries relative to the template. The utmost translation yield was reached at ~5 equiv., leading to ~70% for the 8-mer library (Fig. 2c) and ~30% for the 12-mer library, as confirmed by denaturing polyacrylamide gel electrophoresis (PAGE) (Supplementary Fig. 7c,d). Each artificial FNAP libraries anneal effectively with the cssDNA scaffold, yielding the trivalent combinatorial MEDUSA library (Fig. 2nd, and Supplementary Fig. 7e,f). Resulting from its greater translation yields and to reduce cross-hybridization between longer binding models, we opted to make use of the 8-mer library for affinity choices.
a, Buildings of functionalized and non-functionalized trinucleotide constructing blocks for the manufacturing of the FNAP library. b, The construction of the 8-mer template library with the corresponding Sanger sequencing electrophoretic traces for direct and reverse sequencing runs. c, Translation response of the biotinylated 8-mer template library into the corresponding FNAP library carried out at completely different trinucleotide library concentrations. Non-biotinylated 8-mer library (r8) was loaded as a reference. The FNAP translation product is arrowed. d, Native PAGE evaluation of the meeting of MEDUSA utilizing a synthesized FNAP library. e, Scheme of the multivalent choice cycle for trimeric FNAP assemblies.
Every spherical of choice commences with ligase-mediated translation of the biotinylated template DNA library right into a FNAP library utilizing 5′-phosphorylated trinucleotide constructing blocks. Throughout this course of, the ligase catalyses the templated synthesis of the FNAP area between the initiation and termination primers. Following strand separation and purification, the FNAP library is trimerized utilizing the target-tailored cssDNA scaffold and subjected to affinity choice towards the goal protein, which is immobilized on magnetic beads. For the subsequent choice spherical, the library is warmth eluted and FNAPs are reverse translated again into the DNA template library (Fig. 2e). Because of the target-tailored geometric group, the choice strain is directed in the direction of the enrichment of synergistic multivalent binders. Sequences able to cooperative binding to the goal ought to broaden quicker as a result of their slower dissociation charges. With each spherical, affinity choice based mostly on multivalent interactions turns into extra dominant.
Multivalent affinity choices yield a excessive diploma of sequence range
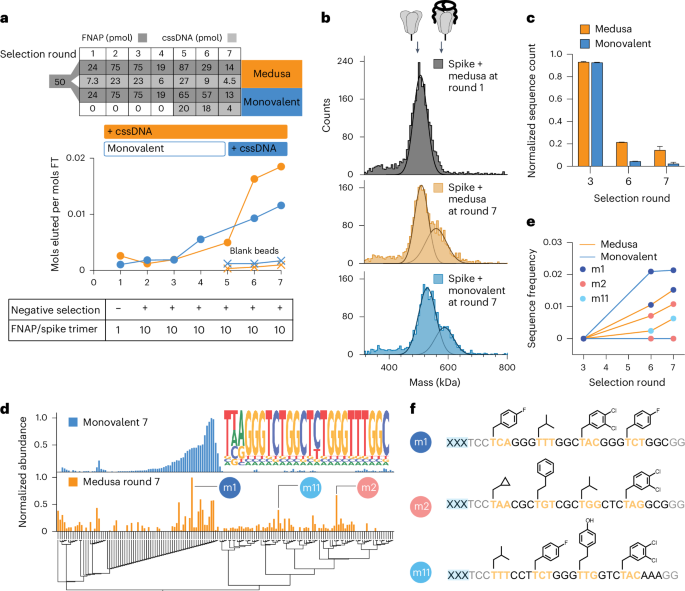
As in SELEX, the frequency of high-affinity sequences within the naïve FNAP library is low, with some binders current in solely a single copy. Furthermore, the distribution of binding affinities is unknown. To deal with the excessive complexity of the naïve library, statistically unfavourable for the stochastic meeting of MEDUSAs throughout early rounds of choice, two choice methods had been examined: (1) completely multivalent and (2) a monovalent pre-enrichment throughout preliminary rounds, adopted by multivalent closing choices. We carried out two parallel choice campaigns towards the SARS-CoV-2 spike protein (Supplementary Fig. 8). The PAGE-purified beginning FNAP library was cut up and one half (termed ‘monovalent’) first underwent 4 rounds of monovalent affinity choices earlier than multimerization. The opposite half was immediately subjected to multivalent choices from spherical 1 (termed ‘medusa’, Fig. 3a, prime). The ratio between the FNAP library and goal was maintained fixed for each methods all through the choice marketing campaign (Fig. 3a, backside). After seven rounds of affinity choice, the binding capability of each the monovalent and MEDUSA libraries started to plateau, prompting us to conclude the choice marketing campaign (Fig. 3a). The presence of binding sequences in each libraries after choice spherical 7 was confirmed by mass photometry (Fig. 3b).
a, Progress in spike-binding choice for multivalent (medusa) and monovalently prefocused (monovalent) choice methods. Bulk affinity of trivalent and monovalent FNAP libraries to trimeric spike protein was assessed by quantifying the quantity of FNAPs within the flow-through (FT) versus elution fractions. b, Enhance in bulk affinity of trimeric FNAP libraries to spike protein by evaluating the MEDUSA assemblies ready with the FNAP library from choice spherical 1 versus choice spherical 7 for each choice methods utilizing mass photometry. c, Development of choice course of indicated by the lower in FNAP library complexity utilizing NGS. Knowledge are introduced as imply ± s.d. (n = 2, unbiased sequencing runs). d, A number of sequence alignment of the highest sequences from NGS information for 2 examined choice methods with corresponding sequence abundances. The frequent motif of sequences retrieved from the monovalent choice course of is displayed as a nested graph. e, Enrichment of three chosen hits over the rounds of choice for 2 examined choice methods utilizing NGS. f, Sequences and side-chain buildings of chosen FNAPs. xxx, scaffold-hybridization area.
Sequencing information at rounds 3, 6 and seven revealed a gradual lower within the variety of distinctive reads within the sequence swimming pools as choice progressed, indicating the enlargement of binding sequences for each choice methods. The range of the ultimate MEDUSA library was round 4.6 occasions greater than the monovalent library, as estimated from the variety of distinctive sequences within the pool (Fig. 3c and Supplementary Fig. 9a,b). We motive two potential processes may contribute to the distinction within the complexities of the libraries: enrichment of synergistically binding sequences that in any other case can not broaden within the monovalent choice regime, and ‘parasitic’ carry-over of low-affinity sequences into the late rounds of choice as a result of their stochastic incorporation into the MEDUSAs composed of high-affinity sequences. As ‘parasitic’ low-affinity sequences successfully render respective MEDUSAs much less lively than the assemblies with greater numbers of cooperatively binding sequences, we motive that the choice is directed in the direction of the lower within the frequencies of low-affinity sequences.
Subsequent-generation sequencing (NGS) information after the ultimate spherical of choice revealed that the sequence pool for monovalent choice technique was ultimately dominated by one household of FNAPs that includes a standard consensus sequence (Fig. 3d, prime). Notably, the found motif will also be present in a number of reported DNA aptamers, thereby additional validating the particular enrichment of binding sequences46 (Supplementary Fig. 10). In distinction, the sequencing information obtained from the medusa library show a extra various vary of sequences along with the aforementioned sequence household (Fig. 3d, backside). The variations in sequence compositions between the 2 libraries are additionally evident within the modifications in side-chain frequencies noticed throughout completely different rounds of choice (Supplementary Fig. 11). To validate the enriched FNAPs, we chosen the three most considerable and sequence-unrelated sequences based mostly on the Levenshtein distances matrix (Supplementary Fig. 12). The sequence m1 represents the household of sequences enriched via monovalent choice, whereas m2 and m11 are the sequences that emerged uniquely via the multivalent, geometry-constrained choice course of (Fig. 3e,f).
Evolution with MEDUSA yields distinctive goal selectivity
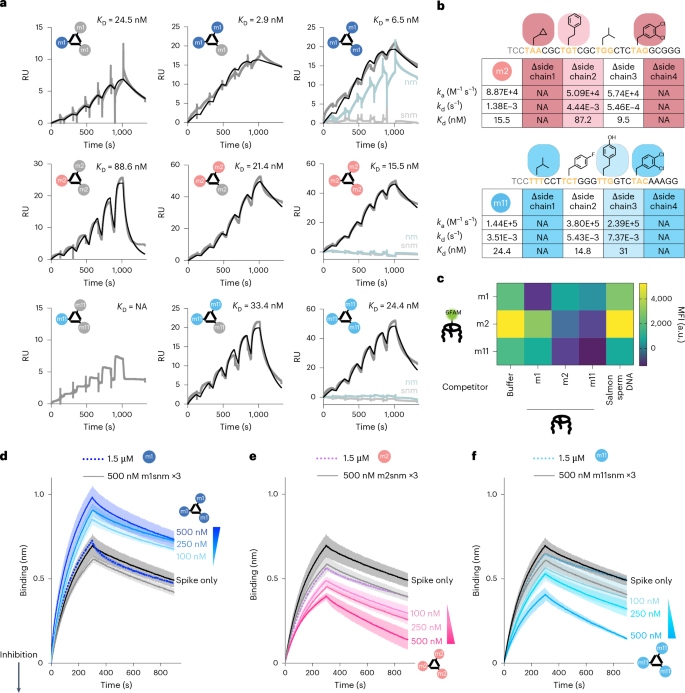
To verify binding affinity for spike protein, the monovalent binding of the full-length and primer-minimized variations of chosen FNAPs (Supplementary Fig. 13a) was examined through floor plasmon resonance (SPR) evaluation. The m1 sequence displayed low-nanomolar monovalent affinity for the spike protein (Supplementary Fig. 13b), and m2 confirmed ~200-nM affinity (Supplementary Fig. 13c). No SPR sign was noticed for monovalent m11, even on the highest analyte focus examined (600 nM). We discovered that the primer area didn’t considerably affect binding efficiency (Supplementary Fig. 13d,e), permitting using primer-minimized variants for additional experiments as a result of greater yields (Supplementary Fig. 14). We ready mono-, di- and trivalent MEDUSAs utilizing the cssDNA multimerization scaffold that includes three orthogonal FNAP-binding websites (cssDNAort), and we additionally ready scrambled and scrambled non-modified management variants. SPR sensorgrams revealed that multimerization of m1 sequence didn’t trigger substantial avidity enhance compared to monovalent m1, suggesting a poor binding cooperativity. In distinction, the rise within the variety of m2 and m11 binding models led to an ∼10-fold enhance in binding power for m2, and a rise from undetectable to 24 nM for m11 (evaluating monovalent and trivalent buildings) (Fig. 4a). This distinction in response upon multimerizations means that m2 and m11 function with completely different binding dynamics, displaying behaviour much like chelation for m2 and cooperative binding for m11 (ref. 47). Utilizing cssDNAort enabled evaluation of all seven doable hetero-FNAP multivalent MEDUSAs (Supplementary Fig. 15a), all displaying comparable nanomolar avidities, with no clearly optimum mixture (Supplementary Fig. 15b,c). Notably, trivalent assemblies demonstrated comparatively gradual affiliation and dissociation charges, which can counsel conformational modifications are required earlier than binding (Supplementary Desk 7), a phenomenon typically reported for aptamer–protein interactions48.
a, SPR sensorgrams characterizing the binding kinetics between trimeric SARS-CoV-2 spike protein immobilized on the CM3 chip and mono-, di- and trivalent assemblies of chosen FNAPs. RU, response models. As controls, assemblies ready with non-modified (nm) and scrambled non-modified (snm) variations of the corresponding binding models had been used. The concentrations of injected assemblies had been 9, 18, 37, 75, 150 and 300 nM. The black curves symbolize the binding kinetics match. b, SPR kinetic parameters (okaya, affiliation price fixed; okayd, dissociation price fixed; Okayd, dissociation fixed) for trivalent supramolecular assemblies ready utilizing side-chain-deficient variants of m2 and m11 sequences. c, Competitors ELISA assay signifies distinct binding specificity between FNAPs chosen through multivalent and monovalent choice methods. The assay was carried out in duplicate (n = 2, technical replicates), and the imply values are plotted. d–f, Competitors BLI sensorgrams depicting spike protein binding to dimeric ACE2-Fc, immobilized on the Protein A BLI probes. Assemblies of chosen FNAPs (d, m1 MEDUSA; e, m2 MEDUSA, f, m11 MEDUSA) had been blended with trimeric SARS-CoV-2 spike protein at three growing meeting concentrations. The lower in mass switch to the BLI probe signifies that the compound interferes with the ACE2–spike protein interplay. Assemblies of scrambled non-modified variants of the chosen sequences had been used as detrimental controls. The gradient triangle signifies the growing focus of FNAP meeting. All measurements had been carried out in duplicate (n = 2, technical replicates), and common indicators had been plotted with the s.d. vary highlighted. NA, not relevant.
Monovalently derived m1 and multivalently derived m2 and m11 sequences displayed dramatically completely different reliance on side-chain modifications for binding to the spike protein. Stripping all modifications from the m1 sequence whereas preserving its DNA sequence (Fig. 4a, ‘nm’) didn’t end in any substantial alteration of the binding affinity. In distinction, removing of modifications from the multivalently chosen m2 and m11 severely disrupted binding. Particular person side-chain contributions in m2 and m11 had been measured by getting ready a scientific set of side-chain-deficient variants and measuring their affinities (Fig. 4b, and Supplementary Fig. 16). Three out of 4 aspect chains had been important for goal binding, a considerably bigger portion in comparison with reported binders with greater functionalization density38. Moreover, NUPACK simulations predict secondary buildings, highlighting the results of high-diversity areas within the choice of binding sequences (Supplementary Figs. 17–19).
Following the proof that multivalent choice yields cooperative binders, we had been curious to research goal selectivity within the context of mutations. Repeating the SPR assays with m1, m2 and m11 MEDUSAs to Omicron BA.4, XBB1.16.1 and Delta B1.167.2 mutants confirmed orthogonal selectivity for m1 versus m2 and m11 (Supplementary Fig. 20). The m1 MEDUSAs had been capable of bind all mutants, hinting towards a scarcity of true goal selectivity however a normal binding interface. Nevertheless, the m2 and m11 MEDUSAs solely certain to the wild-type spike, the goal they had been chosen for, indicating that these binders not solely profit from multivalent cooperativity but additionally present outstanding goal selectivity. We deduce that m1 binds to a distinct bodily part of the spike protein in comparison with m2 and m11, and assessed the binding specificities of the MEDUSAs through competitors ELISA assays. A major lower within the quantity of certain assemblies was noticed for MEDUSAs ready with similar FNAP-binding models. Nevertheless, no competitors was noticed between m1 MEDUSA and MEDUSAs that includes m2 and m11, indicating that geometry-constrained choice certainly reveals sequences concentrating on an orthogonal epitope in comparison with monovalent SELEX. Moreover, binding epitopes of multivalently developed sequences m2 and m11 had been discovered to overlap, as evidenced by diminished FAM fluorescence within the m2–m11 MEDUSA pair (Fig. 4c, and Supplementary Fig. 21). No important competitors was noticed between the chosen MEDUSAs and a subset of reported aptamers with RBD and NTD binding specificities11, suggesting distinct epitopes (Supplementary Fig. 22).
To find out whether or not the noticed binding specificities correlated with useful efficiency variations, we examined their potential to interrogate PPIs between spike and dimeric ACE2 in a aggressive biolayer interferometry (BLI) assay. MEDUSAs displaying m1, m2 and m11 FNAPs had been ready and preincubated with the spike protein. If MEDUSAs intervene with the spike protein–ACE2 interplay, immersing ACE2-functionalized probes into options of spike protein would end in a lower in mass switch to the probe. Nevertheless, the other was noticed with m1 MEDUSA, which confirmed a concentration-dependent enhance in mass switch to the ACE2-coated BLI probe. This means that m1 MEDUSA binding doesn’t create steric hindrance for ACE2 however as a substitute permits your entire MEDUSA–spike protein complicated to bind the probe, growing mass switch relative to the spike protein alone. In distinction, for m2 and m11 MEDUSAs, a marked drop within the BLI sign was noticed, indicating inhibition of ACE2 binding. Notably, this repressive impact was noticed solely with trivalent assemblies and never with the binding models alone, underscoring the significance of multivalency for the useful efficiency of multivalently chosen sequences (Fig. 4d–f).
Scaffold geometry and rigidity interaction for useful exercise
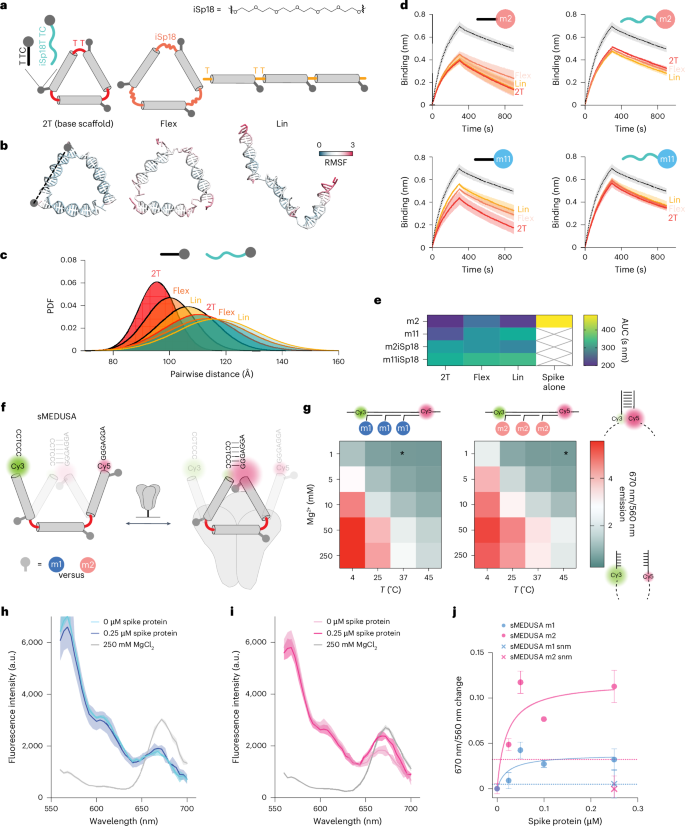
We produced a set of other buildings with elevated conformational flexibility (Fig. 5a, prime) to discover how MEDUSA scaffold and linker configurations affect efficiency. The unique cssDNA scaffold (2T) was linearized (Lin) to discover geometry and we changed one of many T nucleotides by hexaethylene glycol (iSp18) in every of the three vertices to discover flexibility (Flex) (Fig. 5a). Elevated flexibility of those different MEDUSAs was confirmed with OxDNA simulations49 (Fig. 5b,c and Supplementary Fig. 23). Native binding unit flexibility was modified by inserting iSP18 linkers between the m2 and m11 binding models and their scaffold-hybridization areas. The inhibitory potential of all new scaffold and linker variations (Supplementary Fig. 24) was examined in a BLI competitors assay with ACE2 dimer. Our outcomes revealed that modifying the unique geometry and suppleness of ligand presentation diminishes the efficiency of m2 and m11 assemblies. Probably the most notable lower was noticed for assemblies with extreme native linker flexibility (Fig. 5d,e), whatever the scaffold variant. The rise within the meeting’s core flexibility correlated with the lower of the meeting’s inhibitory potential, which was particularly distinguished for the assemblies of the m11 binding unit, which was proven to achieve most from multivalent presentation. We conclude that conformationally constrained scaffolds with target-specific geometry are essential for the inhibitory potential of MEDUSA.
a, A diagram depicting the bottom multimerization scaffold (2T), its linear (Lin) variant and a variant with extra versatile vertex areas achieved by substituting one of many T nucleotides with a hexaethylene glycol spacer (Flex). FNAPs with prolonged spacers separate the binding area from the meeting’s core. Corresponding coarse-grained fashions of the scaffold buildings are given under. b, Shut-to-average oxDNA fashions of the designed scaffolds. RMSF, root imply sq. fluctuation. c, Distribution of end-to-end distances between FNAP attachment factors for varied assemblies, based mostly on coarse-grained simulations. The common distribution of all three distances is plotted for every particle. d, Competitors BLI sensorgrams for FNAP assemblies with varied scaffold and binding unit compositions; spike protein with out assemblies is plotted with a dashed line. All measurements had been carried out in duplicate (n = 2, technical replicates), and common indicators are plotted with s.d. ranges highlighted. e, A heatmap summarizing the imply areas beneath the curve (AUC) calculated from the BLI information for every scaffold and FNAP mixture. f, Schematic of sMEDUSA, which options two conformational states in dynamic equilibrium, with the closed conformation being stabilized upon multivalent cis-interaction with the spike protein (proven in gray). g, Conformational state diagram for m1 and m2 gadgets as a perform of temperature and Mg2+ focus. The situation used within the spike protein binding assays is indicated by an asterisk. h, Fluorescence depth spectral scans for m1 sMEDUSA. i, Fluorescence depth spectral scans for m2 sMEDUSA. The respective spectral scans of sMEDUSAs within the presence of 250 mM MgCl2 are included as a reference for maximal FRET. j, Change in 670 nm/560 nm fluorescence ratio relative to sMEDUSA in buffer for m1 and m2 sMEDUSAs within the presence of accelerating concentrations of spike protein, and for scrambled non-modified (snm) sMEDUSAs at 0.25 μM spike focus. Dashed strains symbolize 670 nm/560 nm ratio change for m1 and m2 sMEDUSAs within the presence of 100 μg ml−1 BSA. All FRET assays had been carried out in triplicate (n = 3, technical replicates), with the typical values plotted and s.d. ranges highlighted.
In an try to translate this optimum geometric binding and goal selectivity into biotechnology instruments, we explored if multivalent target-induced sensors may very well be generated from MEDUSAs. The conformation that facilitates the best ligand positioning ought to be favoured upon the multivalent engagement of its binding models with a multimeric goal. Crucially, this solely holds true if the binding models can interact in synergistic binding in cis. We devised a switchable MEDUSA variant (sMEDUSA) that may undertake two discrete conformational states: a suboptimal open state for multivalent cis-binding, and a extra optimum conformationally constrained closed state. Within the open state, sMEDUSA resembles the Lin construction, whereas within the closed state it’s akin to the unique MEDUSA. The structural foundation for the switchable behaviour of sMEDUSA arises from its scaffold, which incorporates brief (6-nt) complementary areas on the 5′ and three′ termini flanking three FNAP-binding websites (Supplementary Fig. 25). Upon the hybridization of this scaffold with three binding models, a molecular switchable system with dynamic equilibrium between open and closed states is obtained. By introducing a Förster resonance power switch (FRET) pair on the root of the dynamic hairpin, the proportion of closed-state molecules might be monitored in bulk by ratiometric readout of fluorescence intensities at 670 nm (indicative of the closed conformation) and 560 nm (indicative of the open conformation) (Fig. 5f). We efficiently validated the system by modulating the equilibrium between open and closed states utilizing temperature and Mg2+ focus: each a lower in temperature and a rise in Mg2+ focus led to the stabilization of the hairpin and a subsequent enhance within the closed-state inhabitants (Fig. 5g).
To discover the dynamic vary of the system and to check whether or not the identical shift within the equilibrium between the open and closed conformations of sMEDUSA may very well be achieved by cooperative multivalent binding in cis, we carried out a sandbox experiment with sMEDUSAs that includes brief DNA sequences with completely different GC content material rather than FNAP-binding models (Supplementary Fig. 26). We noticed that the conformationally constrained 2T MEDUSA goal was the best in mediating the FRET enhance of the sMEDUSAs, and the rise in GC content material of the binding models correlated with the rise in FRET (Supplementary Fig. 26e). Subsequent, we examined whether or not the conformational dynamics of sMEDUSA may very well be altered by the spike protein (Supplementary Fig. 27). We in contrast FRET efficiencies of m1 and m2 sMEDUSAs, together with sMEDUSAs with scrambled non-modified management sequences. Excitingly, the addition of the protein to the m2 sMEDUSA led to a extra substantial enhance in FRET in contrast with m1 sMEDUSA (Fig. 5h–j), indicating efficient multivalent cis-binding to the spike protein and consequently a shift of the conformational state equilibrium in the direction of the closed conformation. These information lay the muse for the potential of the sMEDUSA system in molecular sensing purposes in addition to non-FRET-based sensor applied sciences.