Supplies
mRNA synthesis
All in vitro transcription mRNA had been synthesized from a PCR template containing a T7 promoter upstream adopted by the codon-optimized open studying body. All constructs include a 5’ UTR, 3’ UTR and 125 polyA tail. All mRNA was transcribed utilizing the HiScribe T7 Excessive Yield RNA Synthesis Equipment (New England Biolabs). Capping was cotranscriptionally carried out utilizing CleanCap Reagent AG (TriLink Biotechnologies). All uridine was changed with N1-methylpseudouridine (TriLink Biotechnologies). In vitro transcription response was handled with DNase to eradicate the template, and dsRNA was eliminated by cellulose clean-up strategies38. The ultimate product was purified by sodium acetate precipitation.
LNP formulation
LNPs had been formulated as beforehand reported with modification8. A lipid combination consisting of Dlin-MC3-DMA or SM102, DSPC (Avanti Polar Lipids), ldl cholesterol (Sigma) and DMG-PEG2000 or DSPE-PEG2000 (Avanti Polar Lipids) was ready in ethanol as a 20-mM inventory. The molar composition used was 50:10:38.5:1.5 molar ratio. The lipid answer was combined by flowing by a microfluidic mixing gadget Nanoassemblr (Precision Nanosystems) with an aqueous mRNA answer in 10-mM citrate buffer (pH 4) at a 1:3 natural to an aqueous quantity ratio at a complete circulation charge of 4 ml per min. The ensuing LNPs had been then diluted twice with PBS (pH 7.4) instantly and additional dialysed in a single day. Then, the following day, LNPs had been filtered by a 0.22-µm filter.
LNP characterization
The dimensions distribution, particle quantity per millilitre and mode measurement of LNP was measured utilizing NanoSight NS300 (Malvern Panalytical). The zeta potential of LNPs was measured utilizing a Zetasizer Nano ZS (Malvern Panalytical). Complete mRNA content material and encapsulation effectivity had been decided by performing a typical RiboGreen (Thermo Fisher) assay.
Molecular cloning of nanobody
Genes encoding the TP1107 sequence had been synthesized as a gene fragment (Built-in DNA Applied sciences) for cloning right into a pET His6 TEV LIC cloning vector23. Plasmids shall be deposited to the Addgene repository.
Single-domain antibody expression and purification
pET-TP1107 was co-transformed alongside pEVOl-pAzF into B-95.ΔA E. coli, which expresses the orthogonal equipment for the incorporation of azPhe in recognition of UAG codon throughout protein translation25. The B-95.ΔA E. coli pressure is a novel expression vector by which 95 of its authentic UAG codons have been changed together with the elimination of launch issue 1 to facilitate the improved incorporation effectivity of azPhe26.
An in a single day tradition was inoculated into contemporary Terrific Broth media with acceptable antibiotics and grown at 37 °C and shaking till the optical density, OD600, reached 0.7–1.0. The one-domain antibody (sdAb) expression was induced by the addition of IPTG (1 mM), l-arabinose (0.02%) and azPhe amino acid (2 mM). Protein expression was continued for an additional 12–14 h at 30 °C earlier than harvesting the micro organism by centrifugation. Bacterial pellets had been harvested by centrifugation (4,000g, 20 min) and resuspended in Ni-NTA wash buffer adopted by cell lysis utilizing a high-pressure homogenizer (Avestin Emulsiflex C5).
On lysis, cell particles was centrifuged (12,000g, 30 min) and the supernatant was collected for purification by way of an immobilized metallic affinity chromatography column. Extra measurement exclusion chromatography was used to take away non-specifically sure proteins utilizing Superdex 75 10/300 GL gel filtration column (GE Healthcare). The sdAb focus was decided utilizing Nanodrop (Thermo) spectrophotometer at 280 nm.
Unfavorable-stain TEM
Unfavorable-stain TEM was carried out by making use of 3 µl of a 0.05 mg ml−1 answer onto a steady carbon TEM grid (EMS 300 mesh), which was pretreated in a plasma chamber (30 s, 15-mA plasma present) adopted by a number of functions of uranyl formate (0.01% w/v). Imaging was carried out on a Thermo L120C TEM gadget at a magnification of 92k, yielding a bodily pixel measurement of 1.55 Å per pixel. Right here 41 pictures had been recorded on a Ceta direct electron detector. Single-particle evaluation was carried out within the RELION v. 3.1.2 software program bundle24. Briefly, pictures had their CTF parameters estimated adopted by automated particle choosing and successive rounds of two-dimensional classification to homogenize the particle stack. This yielded 19k particles for ab initio three-dimensional (3D) mannequin era, which was carried out in cryoSPARC39 and additional 3D refinements had been finalized in RELION, leading to an ~16-Å 3D reconstruction. A Protein Information Financial institution (PDB) mannequin was generated by initially performing rigid-body becoming of the mouse IgG (PDB ID 1IGY (ref. 40)) utilizing UCSF Chimera41. This fitted mannequin then underwent a molecular dynamics versatile becoming refinement utilizing UCSF ChimeraX/ISOLDE42,43 to suit the antibody into the 3D quantity. The resultant PDB was then used as a template for HADDOCK docking44,45,46,47,48 of the nanobody. The docking clusters that finest match the experimental density had been then thought of for an additional spherical of molecular dynamics versatile becoming with tight torsion and distance restraints primarily based on the unique 1IGY mannequin and the beginning mannequin for the nanobody.
Conjugation of TP1107 to DBCO-PEG2000-DSPE
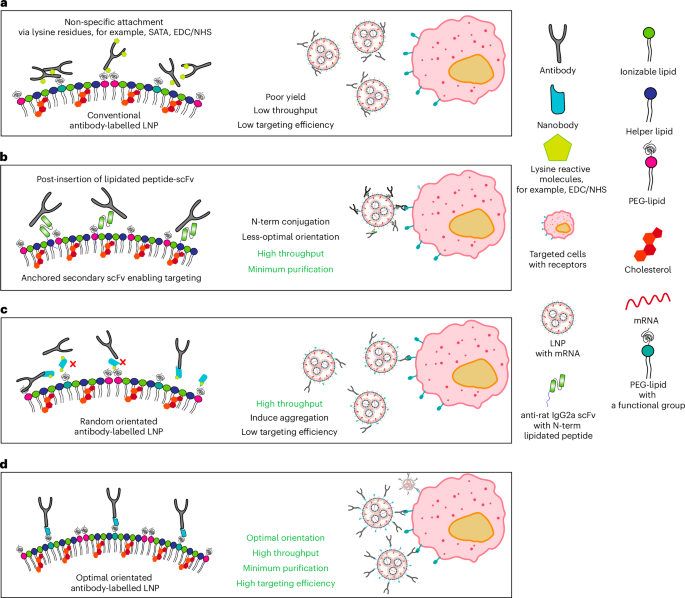
Azide-incorporated TP1107optimum might be instantly conjugated onto DBCO-PEG2000-DSPE by Pressure-Promoted Alkyne-Azide Cycloaddition (SPAAC) chemistry. The conjugation combination was ready at a DBCO:azide molar ratio of two:1. In the meantime, for instance the impact of randomly oriented sdAbs, 2 molar extra of NHS-azide (198 Da, Thermo) was initially conjugated onto TP1107 sdAb. Extra unconjugated NHS-azide linkers had been eliminated utilizing a 7 Okay MWCO Zeba desalting column (Thermo). The azide-modified/azide-incorporated sdAbs had been combined with DSPE-PEG2000-DBCO at a 0.5 molar extra and left for twenty-four h at 37 °C as TP1107random. No additional purification was required.
Publish-insertion of energetic concentrating on module into LNPs and functionalized mAb-TP1107optimum/random LNPs
The post-insertion of DSPE-PEG2000-TP1107 was carried out by including lipidated nanobody to the LNP answer at 0.5% w/w, adopted by temporary mixing earlier than incubating at 4 °C for 48 h unmixed. The free TP1107 or unreacted DSPE-PEG2000-DBCO was eliminated by way of Amicon 100 kDa MWCO (Merck) ultrafiltration. A complete of 5 washes (2,000 rpm, 10 min) had been accomplished. Functionalized LNP was ready by mixing antibodies with TP1107optimum/random LNP on the chosen ratio and incubated 4 °C in a single day.
Calculation of TP1107 per LNP
The sdAb variety of every LNP calculated as beneath:
$${{rm{Quantity}; rm{of}; rm{sdAbs}; rm{per}; rm{LNP}}}=frac{{{rm{Quantity}; rm{of}; rm{sdAbs}; rm{in}; rm{answer}}}}{{{rm{Quantity}; rm{of}; rm{LNPs}; rm{in}; rm{answer}}}},$$
the place the focus of sdAb was calculated from western blot utilizing JESS Easy Western (Bio-Techne) and the variety of LNPs was measured by NanoSight NS300 (Malvern Panalytical).
Concentrating on antibody
Concentrating on antibody used for in vitro and ex vivo research is anti-hTfR (OKT9, bought from WEHI), mouse anti-hCD3 antibody (UCHT1, Thermo Fisher), mouse anti-hCD4 (SK3, BioLegend), mouse anti-hCD5 (UCHT2, Thermo Fisher), mouse anti-hCD7 (124-1D1, Thermo Fisher), mouse anti-hCD22 (eBio4KB128 (4KB128), Thermo Fisher) and mouse IgG1 kappa isotype management (P3.6.2.8.1, Thermo Fisher). Concentrating on antibody used for the in vivo research is mouse anti-mouse CD3ε (QA17A05, BioLegend).
Conjugation of mTfR to DSPE-PEG2000-DBCO
A 5 molar extra of NHS-azide was initially conjugated to purified mAbTfR (OKT9), a sort present from J. Mintern’s group. Extra unconjugated NHS-azide was eliminated by a 7K MWCO Zeba desalting column (Thermo Fisher) following the producer’s protocol. The azide-mTfR was incubated with DSPE-PEG2000-DBCO at 37 °C in a single day at a DBCO:azide ratio of two:1. No additional purification is required.
Preparation of mAblysine LNPs
Roughly 0.05% w/w of mAb-PEG2000-DSPE combination was added to the formulated LNPs. The response was incubated at 4 °C for 48 h. The post-inserted LNP was then concentrated by an ultrafiltration system and slowly utilized to a 90-cm-bed-length gravity-flow measurement exclusion column ready with Sepharose-CL4B gel12,13. The cellular part was PBS. The fractions that contained LNP had been collected and concentrated by the ultrafiltration system. The mRNA focus was measured by a RiboGreen assay, as described earlier than. The particle measurement was decided by NTA.
Cell tradition upkeep
Jurkat cells had been maintained with RPMI media (Gibco) provided with 10% foetal bovine serum (FBS) and penicillin/streptomycin (100 U ml−1). Cells had been cultured at 37 °C in a humidified incubator with 5% atmospheric CO2 together with routine testing or mycoplasma contamination.
Mouse fashions
B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J (IMSR_JAX: 007914) mice had been bought from The Jackson Laboratory and maintained domestically on the Monash animal analysis platform, and mice had been ordered and shipped for the experiment on request. C57BL/6J mice had been obtained from the Monash animal analysis platform. Female and male mice aged 6–14 weeks had been used within the experiments. Ai14 mice had been used for the information proven in Fig. 6 and Supplementary Fig. 14. C57BL/6J mice had been used for the information proven in Supplementary Fig. 14. All of the experimental procedures adopted the protocols authorised by the Institutional Animal Care and Use Committee at Monash College, and the experimental plan was authorised by the Monash Workplace of Analysis Ethics and Integrity Committee beneath ethics approval nos. 37404 and 41587. All of the mice teams had been randomized and gender balanced.
Human PBMC assortment and purification
Wholesome donors aged between 18 years and 50 years previous in each sexes had been recruited voluntarily after the invitation to take part. Ethics is authorised by the Monash College Human Analysis Ethics Committee, software ID 37405. Human blood was collected as per the experimental plan. Right here 10–30 ml of human blood was collected and diluted with PBS earlier than rigorously layered on Ficoll-Paque PLUS density gradient media at 1:1 v/v. A PBMC layer was collected after 400g, 40-min spin and washed with prewarmed RPMI media twice. PBMCs had been both used for experiments or frozen in cell-freezing media at –80 °C.
Antibody-capturing LNP security evaluation and cytokine measurement
C57BL/6 mice had been acquired beneath ethics approval no. 37404. The mice obtained intravenous injections of 0.1 mg kg−1 of assorted LNP formulations or management teams, automobile, unmodified SM102/DSPE LNP, unmodified SM102/DSPE LNP and mCD3 LNP with SM102 and DSPE-PEG2000. Blood samples had been collected at 6 h and 24 h post-injection to judge the impression on liver enzymes (alanine transaminase and aspartate transaminase) and cytokine launch. After 24 h, the animals had been euthanized for liver and spleen harvesting and subsequent histological evaluation. The harvested liver and spleen tissues had been fastened in 10% neutral-buffered formalin for at the very least 48 h. Two mice from every group had been assessed by a veterinary pathologist for additional analysis. Plasma was obtained by centrifuging blood samples at 1,000g for five min at 4 °C and saved as single aliquots. The degrees of mouse cytokines IL-1α, IL-1β, IL-10, IL-6, MIP-1α, MCP-1, IL-2, TNF-α, IFN-γ and IL-4 had been measured utilizing the BD Cytometric Bead Array Mouse Flex Set in accordance with the producer’s protocol. Information assortment was carried out utilizing a Stratedigm S1000EXi circulation cytometer.
Cytokine measurement for the entire blood stimulated with focused LNP
Wholesome donor’s blood was collected on the day of the experiment in heparin-coated assortment tubes. Totally different focused LNPs had been added to every properly at a last focus of two ng μl−1 and incubated for twenty-four h. The blood samples had been then centrifuged to gather plasma for cytokine measurement. Human cytokines IFN-γ, IL-1α, IL-1β and TNF had been quantified utilizing the BD Cytometric Bead Array Human Flex Set in accordance with the producer’s protocol. Information assortment was carried out utilizing a Stratedigm S1000EXi circulation cytometer.
Cell affiliation and transfection assay with functionalized LNPs
To evaluate the binding and transfection efficiencies of functionalized LNPs in Jurkat cells, roughly 50,000 or 100,000 cells had been added to particular person wells in a 96-well plate. A last focus of 0.5 ng µl−1 or 1 ng µl−1 of mRNA was added to the cells and incubated at 37 °C for diverse time intervals. Subsequently, cells had been washed thrice with 2% FBS/PBS following centrifugation at 400g for five min. Cells had been then resuspended in 80 µl of two% FBS/PBS, and the MFI was quantified utilizing a Stratedigm S1000EXi circulation cytometer. eGFP and Cy5 fluorescence had been excited at 488 nm and 642 nm, respectively, with fluorescence emission collected at 520/20 nm and 676/29 nm.
Human PBMC affiliation and transfection assay with functionalized LNPs
To evaluate the binding and transfection efficiencies of the functionalized LNPs, roughly 500,000 PBMCs had been added to particular person wells in a 96-well plate with functionalized LNPs at a last focus of 1 ng µl−1. Then, cells had been incubated at 37 °C for twenty-four h. Then, PBMCs had been washed thrice with 2% FBS/PBS after centrifugation at 400g for five min. To phenotype the subpopulations, cells had been stained towards αCD3-PE mAb (clone OKT3, BioLegend), αCD4-BV510 mAb (clone OKT4, BioLegend), αCD8-BV786 mAb (clone SK1, BioLegend), αCD19-BV421 mAb (clone HIB19, BioLegend), αCD14-Alexa Fluor 700 mAb (clone HCD14, BioLegend), αCD56-BV605 mAb (clone 5.1H11, BioLegend) and viability dye (eBioscience Fixable Viability Dye eFluor 780, Thermo Fisher) on ice for 30 min. All of the antibodies had been used at 1:200 dilutions with Human TruStain FcX (BioLegend) as per the producer’s protocol. After washing away the extreme antibody, cells had been resuspended with 100 µl of two% FBS/PBS for the circulation evaluation (Stratedigm S1000EXi). Cells had been recognized by a mix of floor markers: CD4+ T cells (CD3+ T cells and CD4+ T cells), CD8+ T cells (CD3+ T cells and CD8+ T cells), monocytes (CD3−, CD19−, CD56− and CD14+), NK cells (CD3−, CD19−, CD14− and CD56+) and B cells (CD3− and CD19+). eGFP and Cy5 fluorescence was excited at 488 nm and 642 nm with fluorescence emission collected at 520/20 nm and 676/29 nm, respectively. Information in Supplementary Fig. 8 had been obtained by way of a Cytek Aurora five-laser full spectrum cytometer.
In vivo evaluation of CD3 concentrating on of LNPs to T cells throughout a number of organs
Ai14 mice had been injected intravenously with unmodified LNPs, CD3-targeted LNPs or isotype management LNPs loaded with Cre mRNA. After 24 h, blood was collected by way of cardiac puncture, and the mice underwent transcardiac perfusion with PBS to take away circulating blood. Crimson blood cells had been lysed utilizing ammonium–chloride–potassium buffer (Thermo Fisher) at a 1:10 (v/v) ratio twice, adopted by washing with 2% FBS/PBS. The liver, spleen and lymph nodes (inguinal, iliac and cervical) had been collected and processed as follows.
The liver was minced and digested utilizing a gentleMACS dissociator with 2.8 mg ml−1 of collagenase H and 0.28 mg ml−1 of DNase. The digested combination was filtered to take away the undigested materials and subjected to a gradual spin at 60g. The supernatant was collected and spun down to gather the pellet. The pellet was resuspended in 30% Percoll media and spun to take away hepatocytes, adopted by resuspension with ammonium–chloride–potassium lysis buffer and washing with 2% FBS/Hanks’ balanced salt answer earlier than antibody staining.
The spleen was minced with 1 mg ml−1 of collagenase III and 0.28 mg ml−1 of DNase and digested by fixed, mild mixing till totally digested. Cells had been filtered and pink blood cells had been lysed utilizing an ammonium–chloride–potassium buffer.
Lymph nodes had been collected and homogenized by passing by a 0.45-µm filter. Dissociated cells had been collected and washed with media.
All of the immune cell pellets had been stained with a circulation cytometry panel containing the next antibodies: αCD3e-BV650 mAb (clone 145-2C11, BD Biosciences), αCD90.2-BV650 mAb (clone 53-2.1, BD Biosciences), αCD4-APC-Cy7 mAb (clone GK1.5, BioLegend), αCD8-BV711 mAb (clone 53-6.7, BD Biosciences), αCD19-BV786 mAb (clone 1D3, BD Biosciences), αCD11b-BV421 mAb (clone M1/70, BioLegend), αLy6C-BUV661 mAb (clone HK1.4.rMAb, BD Biosciences), αLy6G-BV605 mAb (clone 1A8, BD Biosciences), αCD45-Pacific Blue mAb (clone S18009F, BioLegend), αI-A/I-E-BV510 mAb (clone M5/114.15.2, BioLegend), αF4/80-PE/Dazzle mAb (clone BM8, BioLegend) and αCD11c-Alexa Fluor 700 mAb (clone N418, BioLegend). Moreover, Mouse BD Fc Block and viability dye (LIVE/DEAD Fixable Blue Lifeless Cell Stain Equipment, Thermo Fisher) had been included. Samples had been incubated on ice for 30 min, adopted by washing to take away extra antibody.
Stream cytometry was carried out utilizing a Cytek Aurora five-laser cytometer, and information had been analysed utilizing FlowJo v10.10.0 (BD Biosciences). Leucocyte phenotyping was carried out utilizing the next markers: CD4+ T cells (CD45+, CD11b−, CD3e+ or CD4+); CD8+ T cells (CD45+, CD11b−, CD3e+ or CD8+); dendritic cells (CD45+, CD3e−, CD19−, CD11c+ or MHCII+); monocytes (CD45+, CD11b+, Ly6C+ or Ly6G−); neutrophils (CD45+ CD11b+, Ly6C+ or Ly6G+); macrophages (CD45+ CD11b+, Ly6C low, Ly6G−, F4/80+ or SSA low); and CD19+ B cells (CD45+, CD11b−, CD3−, CD19+ or MHCII+).
Statistics and reproducibility
Information are introduced as imply ± commonplace deviation (s.d.) primarily based on the information obtained from at the very least n = 3 impartial experiments, wells or mice. Statistical significance was decided utilizing GraphPad Prism 9.0 and said in every determine legend.
Reporting abstract
Additional data on analysis design is offered within the Nature Portfolio Reporting Abstract linked to this text.